About
Summary description:
APECtyper is a free online tool to classify Escherichia coli based on the revised avian pathogenic E. coli (APEC) typing scheme developed by Johnson et al. (2022). The tool utilizes previously-developed typing tools, ECTyper and mlst, along with a custom APEC virulence factor (VF) database (link here). APECtyper outputs include the APEC pathotype classification, individual summary reports from ECTyper and mlst, and a table of BLAST hits to the APEC VF database. A more detailed description of APECtyper’s steps and outputs can be found below in the How APECtyper workssection.
If you use APECtyper in your research, please cite:
-
Johnson TJ, Miller EA, Flores-Figueroa C, Munoz-Aguayo J, Cardona C, Fransen K, Lighty M, Gonder E, Nezworski J,
Haag A, Behl M, Kromm M, Wileman B, Studniski M, Singer RS. Refining the definition of the avian pathogenic
Escherichia coli (APEC) pathotype through inclusion of high-risk clonal groups. Poult Sci. 2022 Oct;101(10):102009.
doi: 10.1016/j.psj.2022.102009. Epub 2022 Jun 14. PMID: 35952599; PMCID: PMC9385700.
https://pubmed.ncbi.nlm.nih.gov/35952599/ -
Johnson TJ, APEC VF Database, GitHub:
https://github.com/JohnsonSingerLab/APEC_VF_database -
Bessonov K, Laing C, Robertson J, Yong I, Ziebell K, Gannon VPJ, Nichani A, Arya G, Nash JHE, Christianson S.
ECTyper: in silico Escherichia coli serotype and species prediction from raw and assembled whole-genome sequence data.
Microb Genom. 2021 Dec;7(12):000728. doi: 10.1099/mgen.0.000728. PMID: 34860150; PMCID: PMC8767331.
https://pubmed.ncbi.nlm.nih.gov/34860150/ -
Seemann T, mlst, GitHub: https://github.com/tseemann/mlst
https://github.com/tseemann/mlst -
Johnson TJ, APEC VF Database, GitHub:
https://github.com/JohnsonSingerLab/APEC_VF_database -
Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M.
Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006 Jun;60(5):1136-51.
doi: 10.1111/j.1365-2958.2006.05172.x. PMID: 16689791; PMCID: PMC1557465.
https://pmc.ncbi.nlm.nih.gov/articles/PMC1557465/
How APECtyper works:
Background
Colibacillosis in poultry is a unique disease manifestation of Escherichia coli in the animal world, as one of the primary routes of entry is via the respiratory tract of birds. Because of this, a novel extraintestinal pathogenic E. coli (ExPEC) subpathotype coined avian pathogenic E. coli (or APEC) has been described. Like other ExPEC, this pathotype has been challenging to clearly define, and in the case of APEC, its role as an opportunistic pathogen has further complicated these challenges.
Many methods have been proposed for typing APEC. A commonly used traditional APEC typing scheme was based on the presence of five genes (iutA, hlyF, iss, iroN, and ompT) found in a pathogenicity-associated island on the ColV and ColBM plasmids (referred to hereafter as APEC plasmids) (Johnson et al. 2008). However, in our 2022 Poultry Science paper, we used whole genome sequencing analysis to show that the APEC plasmid is highly prevalent in both clinical isolates from diseased turkeys and cecal isolates from healthy birds (Johnson et al. 2022). In contrast, we identified distinct differences in clonal backgrounds of dominant turkey clinical isolates versus cecal strains, with a subset of sequence types (STs) in clinical samples (ST23, ST117, ST131, ST355, and ST428), which were rare within cecal samples. Egg lethality assays showed that, irrespective of plasmid carriage, dominant clinical STs are significantly more virulent than dominant cecal STs.
Based on these results, we developed a revised APEC typing scheme that incorporates APEC plasmid carriage plus markers for dominant clinical STs (Figure 1). In this scheme, detection of an isolate belonging to a known high-virulence ST (ST23, ST131, ST355, and ST428) or serogroup O78, classifies it as “high-risk”.
The presence of the APEC plasmid (via possession of at least three APEC genes: iutA, hlyF, iss, iroN, and ompT) with one of the aforementioned markers classifies an isolate as “high-risk APEC”. The presence of the APEC plasmid genes in the absence of high-risk markers does not rule out that an isolate is APEC or virulent, but does not classify it as a high-risk APEC.
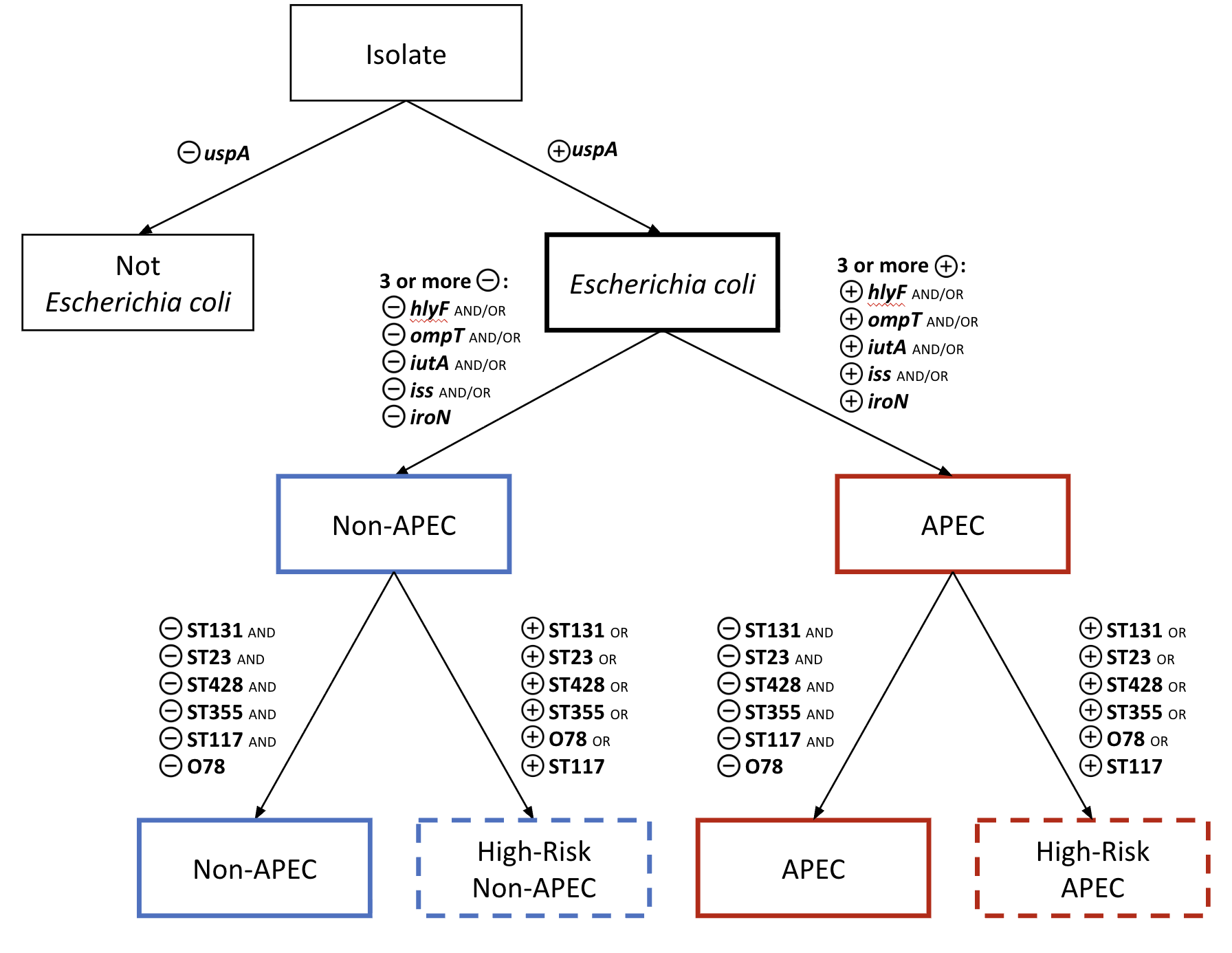
APECtyper steps
APECtyper takes an inputted assembly file (contigs or complete genome) and sequentially runs it through three programs: ECTyper v.2.0.0 (Bessonov et al. 2021), mlst v.2.23.0 (https://github.com/tseemann/mlst), and BLASTX v.2.12.0 (Camacho et al. 2009). Output information from these programs is compiled and processed by a series of custom Python scripts to determine the APEC pathotype classification.
- ECTyper is a speciation and in silico serotype prediction tool. The output includes species designation based on Mash distance (Ondov et al. 2016), matching O- and H-antigen alleles, and quality control (QC) information. If the species is reported as anything other than Escherichia coli, the remainder of the APECtyper pipeline is not run. If the O-antigen is reported as “O78”, the pathotype classification is “high-risk”. ECTyper QC information is provided to allow APECtyper users to make an informed decision about reported pathotype reliability. An explanation of possible QC messages can be found in Table 1 of Bessonov et al. (2021) and the ECTyper GitHub page.
- mlst reports the multilocus sequence type of an assembly based on a designated typing scheme. Here, we use the traditional Achtman seven-gene E. coli MLST scheme (Wirth et al. 2006). Scheme updates are intermittently downloaded from the EnteroBase website (Zhou et al. 2020) to maintain the most complete list of ST profiles. As of July 21, 2025, there were 16,940 unique STs included. If the ST is reported as “ST23”, “ST117”, “ST131”, “ST355”, or “ST428”, the pathotype classification is “high-risk”. Note that it is possible for an assembly to be both a high-risk ST and serogroup O78.
-
A nucleotide BLAST search is performed to identify genes found in the custom
APECvirulence database ,
including the five traditional APEC plasmid genes
(iutA, hlyF, iss, iroN, and ompT) as well as an additional 41 genes associated with APEC.
BLASTn hits are then filtered to those with a percent identity (
pident) ≥ 96% and query coverage per subject (qcovhsp) ≥ 90%. These thresholds were determined based on analysis of APEC gene hits from over 3,700 E. coli isolates from turkey and chicken (dataset from Johnson et al. 2022). Future versions of APECtyper may allow users to specify their own BLASTn percent identity and query coverage thresholds, but the presence of the five traditional APEC plasmid genes for APEC pathotype classification will remain based on the current thresholds. -
A series of custom Python logic scripts merge the outputs from ECTyper, mlst, and BLASTn
to determine the APEC pathotype classification.
- If E. coli is not detected, the pipeline stops and reports “Not Escherichia coli”.
- If at least three of the five traditional APEC plasmid genes (iutA, hlyF, iss, iroN, and ompT) are detected by BLASTn at ≥ 96% identity and ≥ 90% coverage, the assembly file is classified as “APEC”. Otherwise, it is classified as “non-APEC”.
- The assembly file is flagged as “high-risk” if the sequence type (ST) is “ST23”, “ST117”, “ST131”, “ST355”, or “ST428”, or if its O-antigen is serogroup O78.
- The results from the five gene checks are combined with the high-risk assessment to assign one of the four final categories: “APEC”, “High-risk APEC”, “non-APEC”, or “High-risk non-APEC”. If O-antigen or ST information is missing, the classification includes a note that high-risk status is unknown.
References
Bessonov K, Laing C, Robertson J, Yong I, Ziebell K, Gannon VPJ, Nichani A, Arya G, Nash JHE, Christianson S.
ECTyper: in silico Escherichia coli serotype and species prediction from raw and assembled whole-genome sequence data.
Microb Genom. 2021 Dec;7(12):000728. doi: 10.1099/mgen.0.000728. PMID: 34860150; PMCID: PMC8767331.
https://pubmed.ncbi.nlm.nih.gov/34860150/
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL.
BLAST+: architecture and applications. BMC Bioinformatics. 2009 Dec 15;10:421.
doi: 10.1186/1471-2105-10-421. PMID: 20003500; PMCID: PMC2803857.
https://pubmed.ncbi.nlm.nih.gov/20003500/
Johnson TJ, APEC VF Database, GitHub:
https://github.com/JohnsonSingerLab/APEC_VF_database
Johnson TJ, Wannemuehler Y, Doetkott C, Johnson SJ, Rosenberger SC, Nolan LK.
Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool.
J Clin Microbiol. 2008 Dec;46(12):3987-96. doi: 10.1128/JCM.00816-08. Epub 2008 Oct 8. PMID: 18842938; PMCID: PMC2593276.
https://pubmed.ncbi.nlm.nih.gov/18842938/
Johnson TJ, Miller EA, Flores-Figueroa C, Munoz-Aguayo J, Cardona C, Fransen K, Lighty M, Gonder E, Nezworski J,
Haag A, Behl M, Kromm M, Wileman B, Studniski M, Singer RS.
Refining the definition of the avian pathogenic Escherichia coli (APEC) pathotype through inclusion of high-risk clonal groups.
Poult Sci. 2022 Oct;101(10):102009. doi: 10.1016/j.psj.2022.102009. Epub 2022 Jun 14. PMID: 35952599; PMCID: PMC9385700.
https://pubmed.ncbi.nlm.nih.gov/35952599/
Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. Mash: fast genome and metagenome
distance estimation using MinHash. Genome Biol. 2016 Jun 20;17(1):132. doi: 10.1186/s13059-016-0997-x. PMID: 27323842;
PMCID: PMC4915045.
https://pubmed.ncbi.nlm.nih.gov/27323842/
Seemann T, mlst, GitHub:
https://github.com/tseemann/mlst
Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M.
Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006 Jun;60(5):1136-51.
doi: 10.1111/j.1365-2958.2006.05172.x. PMID: 16689791; PMCID: PMC1557465.
https://pmc.ncbi.nlm.nih.gov/articles/PMC1557465/
Zhou Z, Alikhan NF, Mohamed K, Fan Y; Agama Study Group; Achtman M. The EnteroBase user's guide, with case studies on
Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity.
Genome Res. 2020 Jan;30(1):138-152. doi: 10.1101/gr.251678.119. Epub 2019 Dec 6. PMID: 31809257; PMCID: PMC6961584.
https://pubmed.ncbi.nlm.nih.gov/31809257/